Case Study: Sanofi’s Similarity Graph Tool using ChemAxon’s Neo4j Plugin
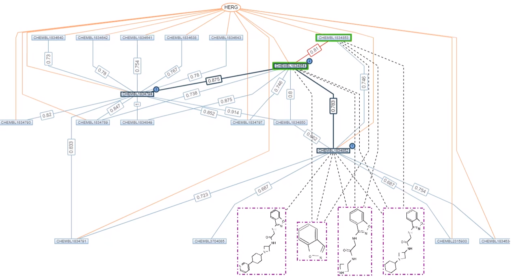
Pic 1: Chemical compounds shown as rectangular nodes (green: matching profile), chemical similarities as blue edges with value, scaffolds colored in dotted pink and projects in orange
JChem Neo4j plugin combines the advantages of the graph database, Neo4j, the high performance and chemical intelligence of ChemAxon's second generation chemical search engine.
The Sanofi Similarity Graph Tool uses these as follows:
- The Neo4j plugin extends the graph database functionality in such a way that chemical structures become searchable, eliminating the need of having them redundant in Oracle
- The structures and corresponding properties are stored within the graph database nodes
- Different relationships are created between nodes.
- It is possible to search and filter on node and relationship properties.
“The Client”
In the validation process for the newest software solution in the ChemAxon (abbreviated CXN below) portfolio, we have spoken with Dan Dragos Stefanescu (abbreviated DDS below) of Sanofi about his experience in the previous 12 months testing the JChem Neo4j Plugin. He combined the Neo4j graph platform with the ChemAxon plugin, and the visualization tool Tom Sawyer Perspectives of Tom Sawyer Software. CXN: Where did your interest in such a graph database plugin come from? “The business need”
DDS: This combination provides an efficient exploration of the chemical space around biologically active chemical matter: ranging from the integration of diverse information linked to compounds; mapping out relationships to related drugs and commercially available compounds. The navigation (traversing) feature and visualization facilitate the exploitation of neighborhood relationships of the different compounds.
We were seeking answers to questions like:
- What are the nearest neighbors to a given compound A that contain scaffold A and show a high permeability?
- Which compounds show activities on targets A and B and have a reasonable ADME profile?
- Is there a commercially available compound similar to compound A that comes with pharmacological data that might be used as a tool compound?
“Highly responsive and user-friendly tools”
CXN: What were the most attractive qualities in this setup for you?
DDS: This integration serves us well in opening avenues for an interactive and visual data traversing of the chemical space. The JChem Neo4j plugin was tested by ChemAxon on a database consisting of 100 million structures. The current project at Sanofi uses millions of structures. Our Neo4j graph database, along with the JChem plugin delivers broad chemical search performance to retrieve data from large data sets. It eliminates the need for performing an additional chemical structure search in Oracle. We have discovered possibilities in new high-end visualization to depict complex relationships that might otherwise remain hidden. The graph database facilitates adding new relationships and node types without compromising the existing data model.
CXN: What features of your similarity graph tool do you find most innovative?
DDS: It retrieves nearest neighbors of a molecule, highlighting highest, second highest (…) chemical similarity edge of a molecule node for interactive graph traversal. It also allows scientists to track the path and order of visited compounds. There is an export function for selected compound IDs for further analysis in other tools. Along with filtering on edge and node properties, it is also possible to apply color-coding rules to molecule nodes. The similarity graph tool finds the shortest path between molecules with respect to the biological context, considering visible nodes either of the currently displayed graph or the entire database. It also allows for the display of scaffolds, finding and highlighting similarities between compounds. Nodes might be enriched with additional data from CSV files, for instance by linking based on compound ID. This data can also be used for color coding and filtering. Furthermore, this web application supports the retrieval of compound similarities based on a common biological function.
In previously used solutions the data were only stored in relational databases, which could considerably slow down even a single nearest neighbor search, with a compound collection walk-through requiring a series of complex searches that might take hours.
After the confirmation and fine tuning of the solution with Sanofi and Tom Sawyer Software, we are proud to release the newest member of the ChemAxon JChem technology software suite.
For further information visit: http://bit.ly/2KGOret
All examples shown are based on publicly available CHEMBL data and for illustration purposes only.