Cheminfo Stories presents: Design Day reviewed by Wendy Warr
Designing New Molecules
Exploring activity cliffs using graph databases
Jan Christopherson of ChemAxon presented some of his work exploring matched molecular pairs (MMPs),2 and in particular the concept of activity cliffs,3 using ChemAxon tools in graph databases. The MMPs were generated using the ChemAxon JChem Extensions in KNIME. Neo4j was the primary interface used to interact with the graph networks generated in the analysis. Visualization was carried out with Cytoscape, plus extension applications named “chemviz2” and “Cypher queries” in order to generate visualization with chemical structures depicted. Cypher is Neo4j’s graph query language. The data studied were pIC50 values for JNK1 and JNK2 inhibitors from CHEMBL. While graph databases may be limited relative to relational database systems when it comes to certain aspects of scaling, they are excellent for exploring data that are highly related. ChemAxon have developed a proof-of-concept search cartridge that interfaces with Neo4j and provides chemical searchability. The highly relational nature of matched molecular pairs makes them a natural fit for representation in a graph database. In one image Jan showed a set of activity cliffs detected by simply assessing the relationships’ activity values. He then used the features of a graph database to explore more fluidly the space around the activity cliff, and by carrying out chemical similarity searches. Having identified an MMP that leads to activity cliff behavior, he explored around the cliff. Jan also did some work on activity pairs3 but his limited dataset did not reveal any concrete examples. While a reaction similarity search has not yet been implemented in the Neo4j cartridge, a good workaround is to set up a separate graph database, using the line graph of the original database. The same was done to the dataset by creating nodes that represent the transformations. If Jan found an interesting transformation he could run a substructure search with this transformation as the query, to find other similar queries of interest. In a typical MMP database, you have to run all the searches first and then create a graph based on the results. Jan’s methodology can create a general MMP analysis. You can carry out simple exploration of related chemical entities, and add multiple activities and activity changes in one or more relationships. You could also add Structure Activity Landscape Index (SALI) relationships.
Marvin Live: the collaborative design platform at UCB
UCB (Union Chimique Belge) is a global biopharmaceutical company focused on transforming the lives of people living with severe diseases in immunology and neurology. UCB employs more than 7,600 people in 40 countries. More than 25% of the company’ revenues are plowed back into R&D. There are two main research centers, in Slough, United Kingdom and Braine-l’Alleud, Belgium. UCB also acquired three sites in the United States and one in London, United Kingdom. Collaborative work across all these sites and time zones is a challenge.
Judi Neuss, an IT specialist at Slough, and Karine Poullennec, a medicinal chemist at Braine-l’Alleud described ChemAxon’s role in a solution addressing this challenge. Between 2014 and 2017, UCB used ChemAxon’s Compound Registration and JChem Engines. In 2018-2019, the two companies embarked upon a new collaboration around the design-make-test-analyze (DMTA) cycle. At UCB the “hypothesis” is a key to the cycle. Once a hypothesis has been established, new ideas to address it are tested. Molecules are then assessed for progression, before prioritization for synthesis. Tracking and biological testing follow. The data are then analyzed, closing the cycle.
UCB required a live discovery platform to enable scientists to share hypotheses and together create ideas across teams located in different time zones. Marvin Live was the preferred solution for a number of reasons. It has a simple and intuitive interface; strong underlying chemistry, and the ability to integrate a virtual registration system; a good selection of plugins and web services which could be configured to suit UCB; nice collaboration features; and a realistic price. A few features UCB required were missing in the version ChemAxon originally demonstrated but Marvin Live was evolving fast and UCB could take advantage of new product developments.
So, UCB committed to running a six-month pilot project, beginning in January 2019. The dual aims were to assess the value of using a collaborative design platform for real projects, for a period long enough to cover several design cycles; and to initiate improvements in cross-team, cross-site, and cross-functional collaboration. The team for the trial consisted of 22 people (medicinal chemists, computational chemists, biologists, ADME specialists, and structural biologists) in two research project teams. Touchscreens were installed in the meeting rooms to enhance the collaborative design experience. At the end of the pilot project, the participants were asked for feedback through a survey and the results were used to evaluate Marvin Live against success criteria.
UCB configured a virtual registration system for assigning virtual compounds IDs. They set up idea properties (project, hypothesis, author, status, priority etc.); plugins and web services (structure checker, preferred property calculation, regulatory status check, submission for docking, search of the virtual registry system, and retrieval of assay data); an Oracle database; and LDAP authentication. New product developments were table view; an editable view to show the key challenge that the ideas in a room were designed to solve; and new functionalities to make Marvin Live work better with touchscreens. Table view is a tabular overview to track design status and prioritization as ideas are progressed to synthesis.
There are many ways a scientist can use Marvin Live. The “one project, one public room per chemical series” is the only idea repository: Excel is not allowed. This is used and updated daily. There are regular dedicated Marvin Live design meetings where ideas are discussed and prioritized.
Centralization and the one-stop shop were liked. Over 90% of users thought that Marvin Live was easy to use; 60% found it useful. Other positive feedback was that Marvin Live enables everyone, even juniors, to contribute to designs, and the use of Marvin Live triggers discussion. On the negative side, users said that it was time-consuming to update rooms, table view was not flexible or customizable; filter functions were limited; and Marvin Live does not capture generic ideas, and does not have enough links to assay data.
On balance, the steering group decided that the pilot project was a great success. All features implemented in the pilot project were moved into production, and more ADME and local mode predictions were added. Master Data Management (MDM) was used for discovery project names.
There are some bugs, and it is still slow to update the rooms. UCB would like better ways to record the hypothesis and to be able to attach files to the hypothesis. They want some improvements to make the table more like Excel (e.g., better sorting and filtering, and the ability to move the columns around). They would also like more integration with data analysis, molecular modeling, and other software; global search; and a tree map to follow idea devolution to see if the idea worked.
Nevertheless, Marvin Live has created new ways of working in UCB; the numbers of users and of designs have increased; and there is an increasing “ping pong” of ideas. The system improves collaboration, transparency, and progression of ideas.
Workshop on Design Hub
Marvin Live has evolved into ChemAxon’s Design Hub. A two-hour workshop focused on how solutions in the new Design Hub can be used in lead optimization and compound tracking. Dóra Barna of ChemAxon gave an overview of the application. In the hypothesis part of the DMTA cycle, scientists need to analyze previous cycle data, prioritize compounds using ideas from the literature, check the status of a project, find out why a certain compound was made and what was learned, trace elusive files, and so on. Marvin Live already featured molecule design but Design Hub adds many more facilities (Figure 3). One is handling the hypothesis: all the evidence supporting an idea (the scientific rationale behind it). Plugins for docking and clustering are new. Teams, projects, registration, status tracking, and assignment have been added. Commenting, file attachments, and tags are new. Universal search and role-based access control are other added features.
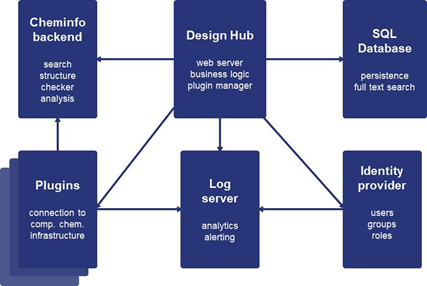
Figure 3. Design Hub technology.
The design history from legacy projects can be searched based on description, keywords, and structural information. A hypothesis is made from the scientific rationale and tested by the design of compounds. For the plugins, NodeJS and Java starting kits, a Python helper library, and serverless support (AWS Lambda and AWS Fargate) are available. More than 50 examples have been published, with source code on GitHub. Searches of eMolecules can be carried out. The hERG assistant has a Matched Molecular Pairs based knowledge base. The model is trained on data from a patch clamp assay in ChEMBL. There is a REST API to JChem Base. The hERG assistant will soon be extended with the new ChemAxon hERG predictor. There is also a REST API to Compliance Checker. An open source docking engine, with a REST API to ChemAxon property predictors, produces a protein-ligand interaction display. You can also use your own docking engine. Enamine REAL substructure and (very fast) similarity searches are carried out with a REST API to JChem Microservices. Soon there will be an “all public database search as a service”. After design, compounds are prioritized for synthesis. The progress of projects can be followed. Design Hub has features for projects, virtual registration, graphical hypotheses, virtual and real data, and compound grouping, and kanban (a framework used to implement agile software development) for productivity. Design Hub Basic and Professional Subscriptions are cloud-only and vary in the number of plugins and users allowed. Design Hub Enterprise is on-premises or hosted.